Note
Click here to download the full example code
Explaining Local Classifier Per Parent Node
A minimalist example showing how to use HiClass Explainer to obtain SHAP values of LCPPN model. A detailed summary of the Explainer class has been given at Algorithms Overview Section for Hierarchical Explainability. SHAP values are calculated based on a synthetic platypus diseases dataset that can be downloaded here.
Out:
<xarray.Dataset>
Dimensions: (class: 15, level: 3, sample: 246, feature: 9)
Coordinates:
* class (class) <U16 'Allergy' 'Bee Allergy' ... 'Respiratory'
* level (level) int64 0 1 2
Dimensions without coordinates: sample, feature
Data variables:
node (sample, level) object 'hiclass::root' ... 'Food Allergy'
predicted_class (sample, level) object 'Respiratory' ... 'Milk Allergy'
predict_proba (sample, level, class) float64 0.16 nan nan ... nan nan nan
classes (sample, level, class) object 'Allergy' nan nan ... nan nan
shap_values (level, class, sample, feature) float64 0.01101 ... nan
from sklearn.ensemble import RandomForestClassifier
from hiclass import LocalClassifierPerParentNode, Explainer
import shap
from hiclass.datasets import load_platypus
# Load train and test splits
X_train, X_test, Y_train, Y_test = load_platypus()
# Use random forest classifiers for every node
rfc = RandomForestClassifier()
classifier = LocalClassifierPerParentNode(
local_classifier=rfc, replace_classifiers=False
)
# Train local classifier per parent node
classifier.fit(X_train, Y_train)
# Define Explainer
explainer = Explainer(classifier, data=X_train.values, mode="tree")
explanations = explainer.explain(X_test.values)
print(explanations)
# Filter samples which only predicted "Respiratory" at first level
respiratory_idx = classifier.predict(X_test)[:, 0] == "Respiratory"
# Specify additional filters to obtain only level 0
shap_filter = {"level": 0, "class": "Respiratory", "sample": respiratory_idx}
# Use .sel() method to apply the filter and obtain filtered results
shap_val_respiratory = explanations.sel(shap_filter)
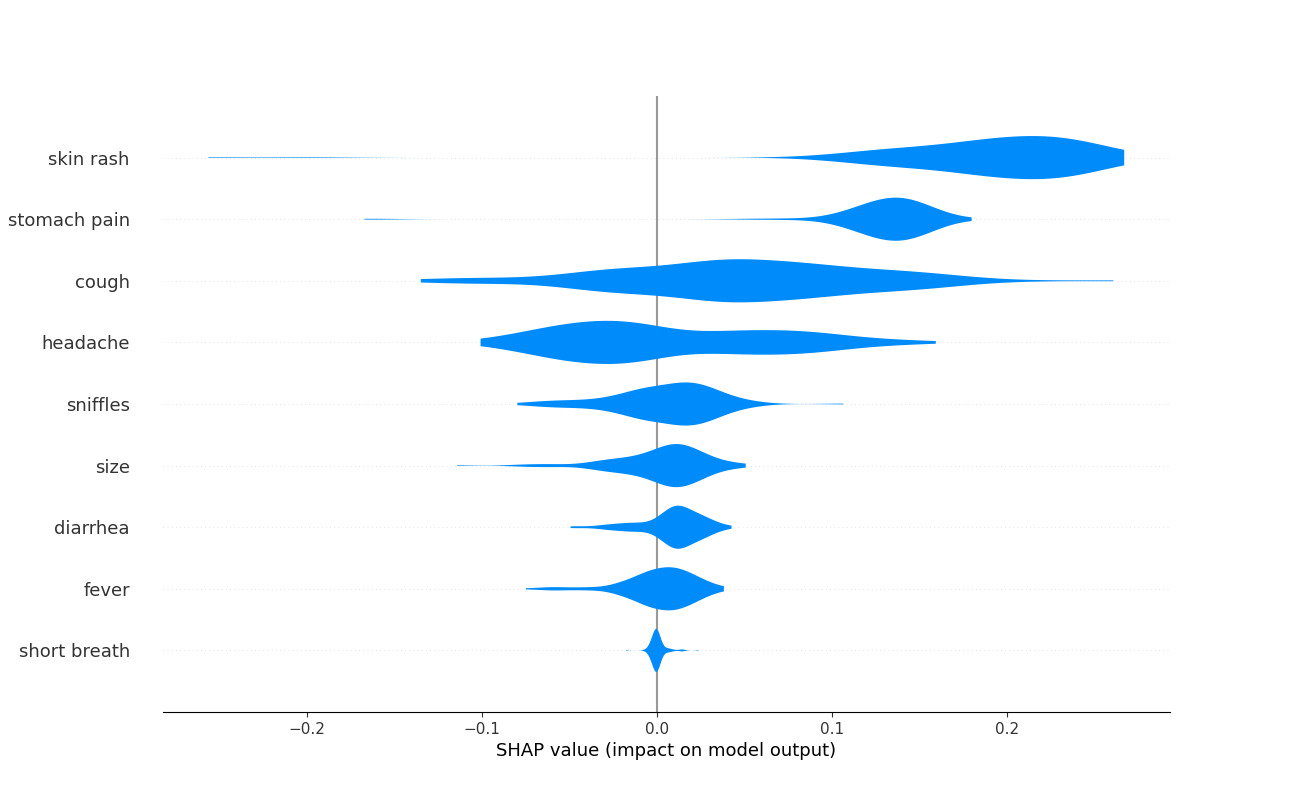
# Plot feature importance on test set
shap.plots.violin(
shap_val_respiratory.shap_values,
feature_names=X_train.columns.values,
plot_size=(13, 8),
)
Total running time of the script: ( 0 minutes 34.983 seconds)