Multi-Label Classification
HiClass supports hierarchical multi-label classification. This means a sample can belong to multiple classes at the same hierarchy level.
On this page, we motivate, explain, and demonstrate how hierarchical multi-label classification is implemented in HiClass.
Motivation
In numerous hierarchical classification problems, it is possible for a sample to be associated with multiple classes at the same level of the hierarchy. This occurs when the classes are not mutually exclusive. For instance, let us consider a problem involving the classification of dog breeds, where we aim to determine a dog’s breed based on available data. Without allowing for multiple paths through the dog breed hierarchy, we would have to assign a single label to each sample, which means we have to choose a single path through the hierarchy, assigning a dog to a single breed. However, this only sometimes reflects reality since a dog can be a mix of multiple breeds. For example, a dog can be a mix of a Dachshund and a Golden Retriever. In such a scenario, we aim to assign both the Dachshund and Golden Retriever labels to the sample, which requires at least two paths through the hierarchy. The following figure illustrates this example.
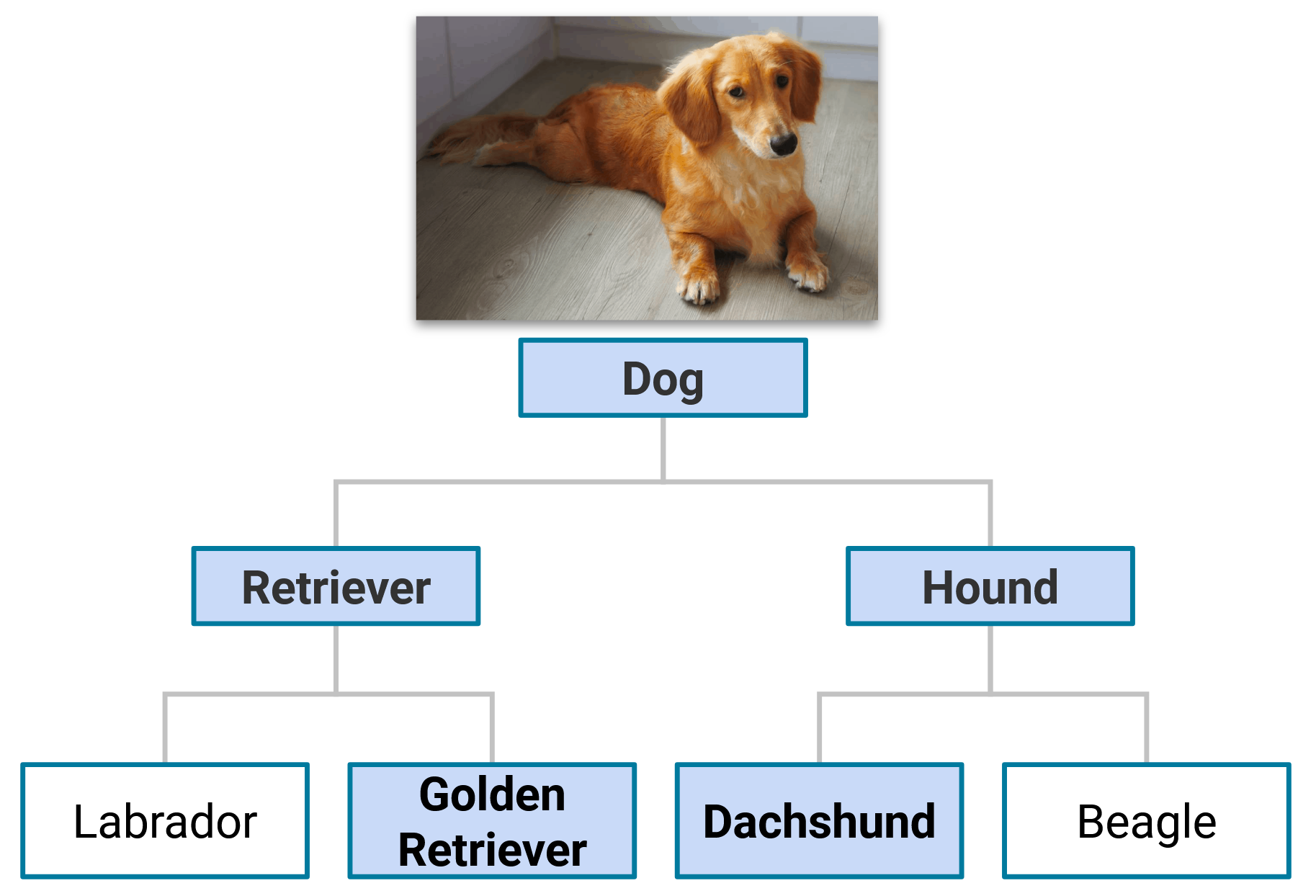
An example image of a dog that is a mix of a Dachshund and a Golden Retriever, thereby requiring multiple paths through the hierarchy for correct classification.
Another multi-label classification example is document classification, in which we aim to classify a document based on its content. The categories are often hierarchical in nature, such as classifying documents into broad topics like “Technology”, “Sports”, and “Politics”, which further have subcategories like “Artificial Intelligence”, “Football”, and “International Relations”. A document can belong to multiple categories, for example, a text that deals with the influence of advancements in AI on International Relations, which can only be correctly classified by multiple paths through the hierarchy.
Background - Classification Terminology
To explain what we mean by hierarchical multi-label classification, we first need to define some terminology.
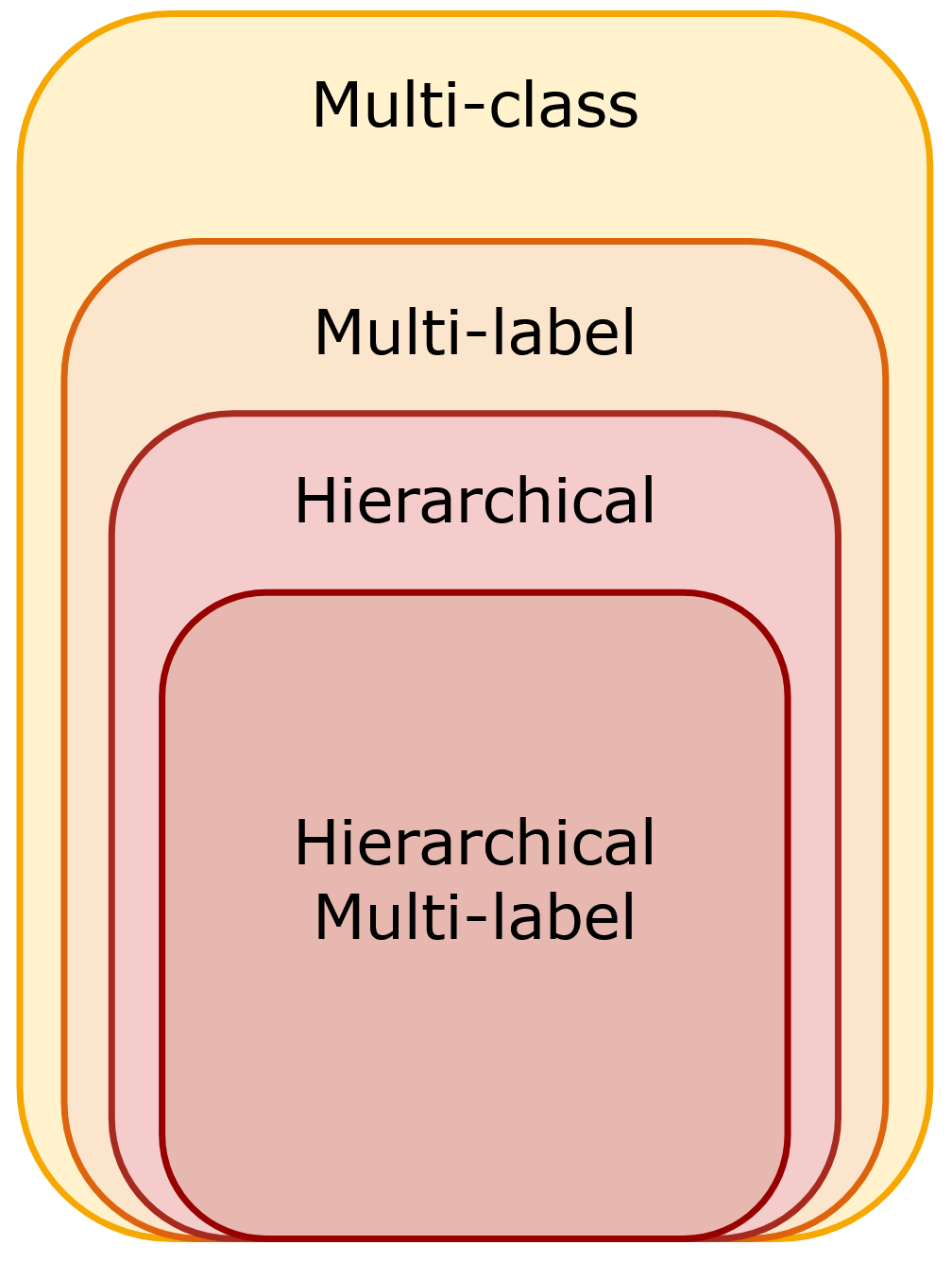
The set of classification problems from most generic (multi-class) to most specific (hierarchical multi-label classification).
In a multi-class classification problem, a sample can be assigned to one class among several options. In a multi-label classification problem, a sample can be associated with multiple classes simultaneously. A hierarchical classification problem is a type of multi-label classification problem where classes are organized in a hierarchical structure represented as a graph, such as a tree or directed acyclic graph (DAG). In this graph, the nodes correspond to the classes to be predicted. If not specified, it is usually assumed that a sample can only belong to one class at each level of the hierarchy. This means a sample can only be associated with a single path through the hierarchy, starting from the root node and ending at a leaf node. In hierarchical multi-label classification, this restriction is lifted. A sample can belong to multiple classes at any level of the hierarchy, i.e., a sample can be classified by multiple paths through the hierarchy.
Design - Target Format
HiClass is designed to be compatible with the scikit-learn API. For the non-multi-label hierarchical classification case, the target array follows the sklearn format for a multi-label classification problem. However, since there is no sklearn specific multi-label hierarchical format, HiClass implements its own format extension. The HiClass target format extends the non-multi-label hierarchical classification format by adding a new dimension to the 2-dimensional array, which captures the different paths through the hierarchy.
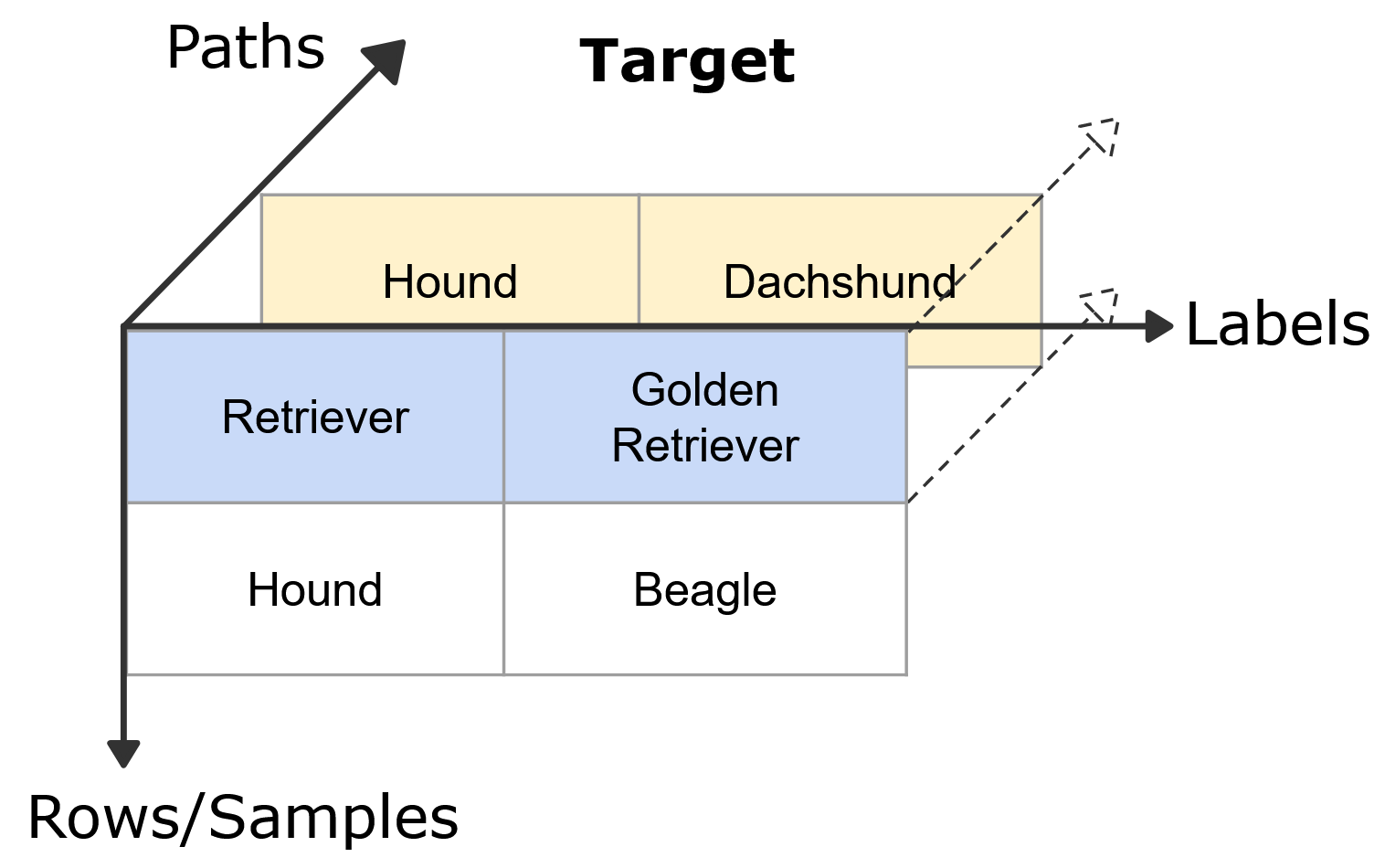
HiClass hierarchical multi-label classification format extension for samples classified by the dog breed hierarchy.
This is implemented as a nested list of lists, in which the last dimension specifies a path through the hierarchy.
y = [
[["Retriever", "Golden Retriever"], ["Hound", "Dachshund"]], # sample 1
[["Hound", "Beagle"]] # sample 2
]
Important to note here is that we specify the whole list of nodes from the root to the most specific nodes for each path.
Even in cases where only the leaf nodes are different, we still need to specify the whole path.
For example, if sample 1 belonged to the Labrador class instead of the Dachshund class, we still need to specify the whole path from the root to the Golden Retriever and Labrador nodes, which would be [["Retriever", "Golden Retriever"], ["Retriever", "Labrador"]].
This is a consequence of using Numpy arrays for the implementation which require fixed dimensions for the target array.
Furthermore, by explicitly specifying the whole path from the root to the leaf node, the target format is readable and easy to comprehend and also works well for hierarchies that are not trees but DAGs.
Fitting the Classifiers
In this section, we outline how fitting of the local classifiers is implemented in HiClass for hierarchical multi-label classification.
Here, we only focus on the hierarchical multi-label classification case for the hiclass.MultiLabelLocalClassifierPerNode and hiclass.MultiLabelLocalClassifierPerParentNode classifiers.
For a recap on how the strategies work, visit the Algorithms section.
Local Classifier Per Node
The hiclass.MultiLabelLocalClassifierPerNode strategy fits a binary local classifier for each node in the hierarchy.
hiclass.BinaryPolicy defines which samples belong to the positive and which ones to the negative class for a given local classifier.
HiClass implements that positive and negative samples for a local classifier are mutually exclusive, i.e., a sample can only belong to a local classifier’s positive or negative class.
In the hierarchical multi-label case, a sample belongs to the positive class if it belongs to any of the paths through the hierarchy that are associated with the local classifier.
For instance, the example image is assigned to the positive class for the Retriever classifier since it belongs to the Golden Retriever class, which is a child of the Retriever node. It is also assigned to the positive class for the Hound classifier since it does not belong to the Dachshund class, which is a child of the Hound node.
Local Classifier Per Parent Node
The hiclass.MultiLabelLocalClassifierPerParentNode trains a multi-class classifier for each non-leaf/parent node, i.e., a node with children in the hierarchy.
The classes to be predicted are the children of the node.
For the multi-label case, a sample can belong to multiple children of a node.
Internally, this is implemented by duplicating the sample and assigning each duplicate to one of the node’s children.
The classifier does not need to support the sklearn multi-label format and can be a standard sklearn classifier.
Prediction
So far, we have only discussed the fitting of the classifiers; in this section, we outline how the prediction is implemented in HiClass for multiple paths. HiClass follows a top-down prediction strategy in which a data sample is classified by nodes in the hierarchy, starting from the root and going down to the leaf nodes. In the single path case, the data sample is assigned the label with the highest probability at each level. This leads to only a single path through the hierarchy for each data sample.
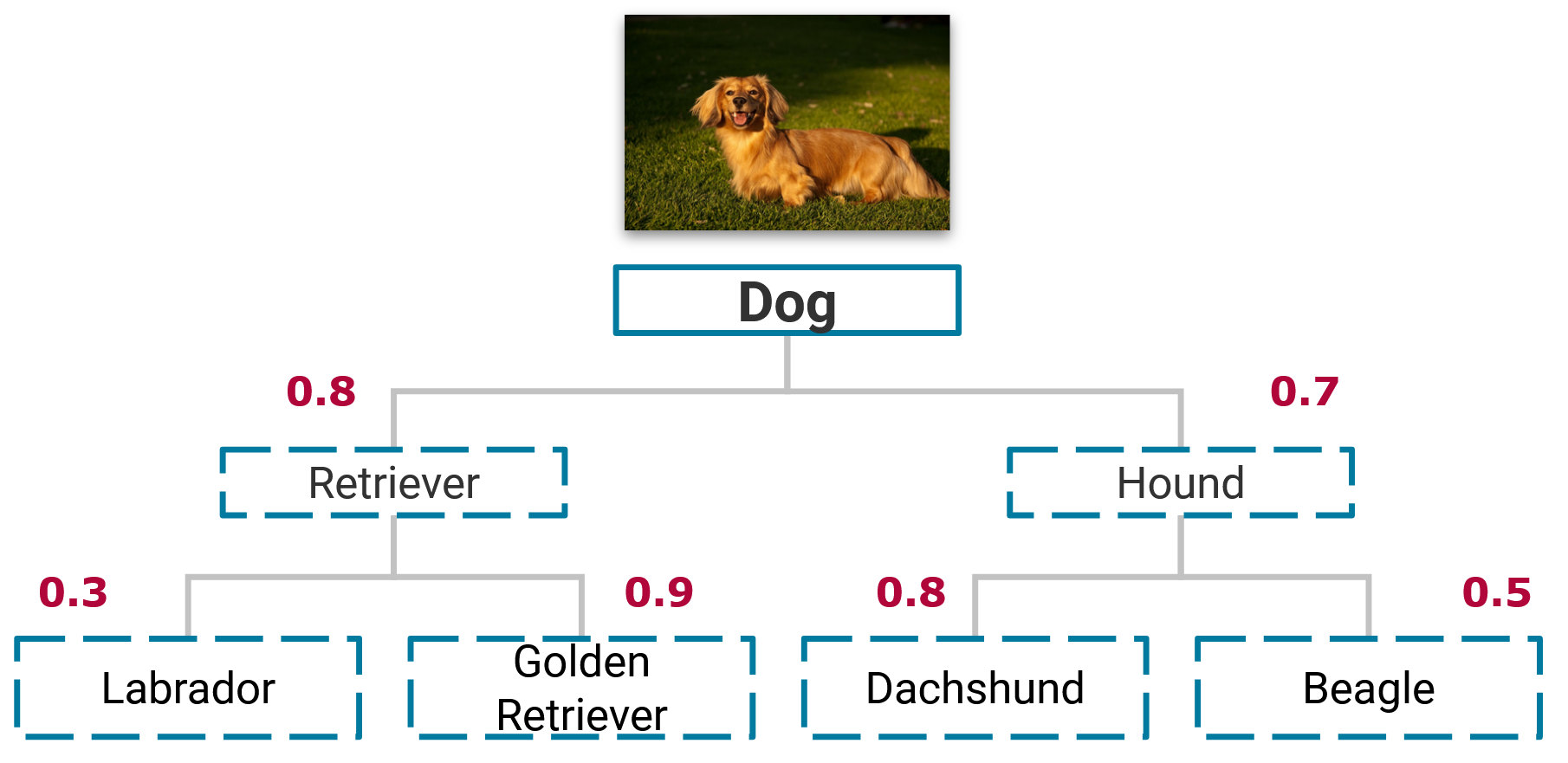
Predicting the labels for a sample using the top-down prediction strategy. Numeric values in red are the predicted probabilities for each node.
In the example given above, the sample would be assigned the label ["Retriever", "Golden Retriever"], since this is the path with the highest probability starting at the root node.
In contrast, when we want to allow for multiple paths through the hierarchy, we need to specify a criterion different from taking the highest probability to assign labels to data samples.
HiClass implements two strategies for this: Threshold and Tolerance.
Threshold
The Threshold strategy assigns a label to a data sample if the probability of the label is above a given threshold. The threshold \(\lambda \in [0, 1]\) is a parameter passed to the predict function and specifies an absolute probability value.
In the example given above, if we set \(\lambda = 0.6\), we would assign the label [["Retriever", "Golden Retriever"], ["Hound", "Dachshund"]] to the sample since the probabilities of the assigned nodes are greater than 0.6.
While this strategy is simple to implement and understand, it has the disadvantage that it is impossible to specify a different threshold for each node in the hierarchy, requiring a global threshold for all nodes.
Furthermore, with the top-down prediction strategy, if the predicted probability is below the threshold for a node, the prediction stops regardless of the probabilities of the nodes further down the hierarchy.
For example, if \(\lambda = 0.85\), no label is assigned to the sample since the probabilities for the Retriever and Hound class are below the threshold value and traversing the hierarchy stops.
Tolerance
The Tolerance strategy mitigates the problem that arises from the absolute probability value in the Threshold strategy by assigning a label to a data sample if the probability is within a given tolerance of the highest probability for neighboring nodes. The tolerance \(\gamma \in [0, 1]\) is a parameter that is passed to the predict function and specifies a relative probability value.
This strategy has the advantage of always predicting at least one class at each level since the tolerance is relative to the highest probability.
For example, with \(\gamma = 0.3\) we would predict the labels [["Retriever", "Golden Retriever"], ["Hound", "Dachshund"], ["Hound", "Beagle"]].
Note that the Beagle label is assigned in the second level because its probability of 0.5 is within the threshold of 0.3 of the highest probability of 0.8 (Dachshund class) of a neighboring node.
Metrics
We extend the hierarchical precision, recall, and F-Score metrics to evaluate the performance of the hierarchical multi-label classifiers. The hierarchical precision, recall, and F-Score are defined as follows and are also defined in Metrics.
Here, we give an example of the hierarchical precision and recall for the multi-label case.
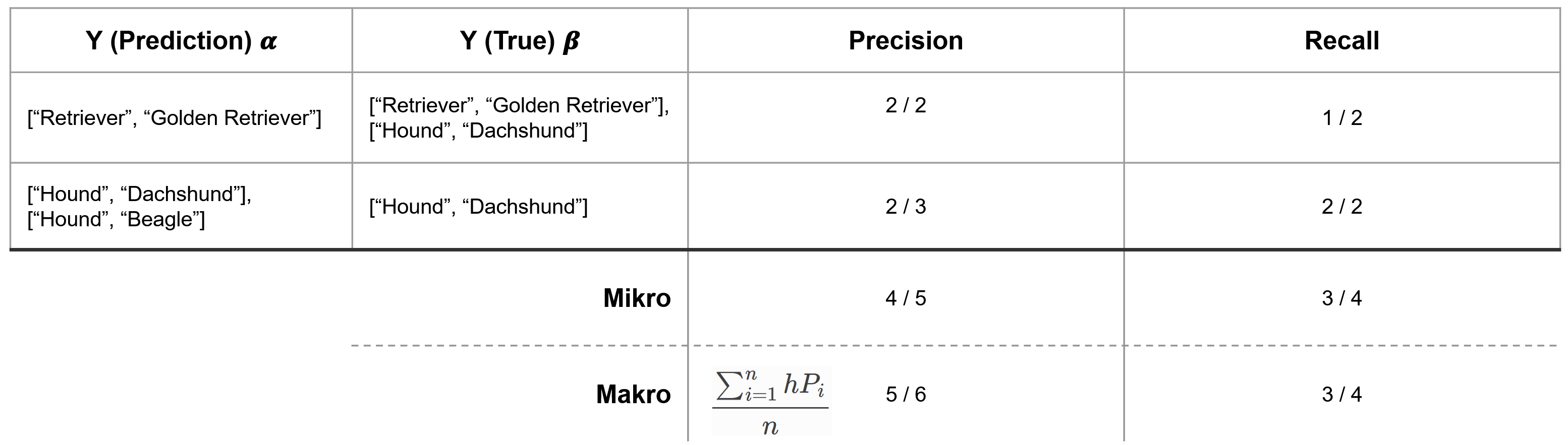
Note that we can define micro and macro averages when calculating the hierarchical precision and recall for multiple samples. The micro-precision/recall of all predictions are considered together, regardless of the sample. In contrast, in the macro precision/recall, we first calculate a sample’s hierarchical precision/recall and then aggregate the results. Since samples can have differing numbers of labels assigned to them, micro and macro averages can lead to different results.
Code example - Putting it all together
Out:
[[['Retriever' 'Golden Retriever']
['Hound' 'Dachshund']]
[['Retriever' 'Golden Retriever']
['' '']]
[['Hound' 'Dachshund']
['Hound' 'Beagle']]]
from sklearn.tree import DecisionTreeClassifier
from hiclass.MultiLabelLocalClassifierPerNode import MultiLabelLocalClassifierPerNode
# Define data
X_train = [[1, 2], [3, 4], [5, 6]]
X_test = [[1, 2], [3, 4], [5, 6]]
# Define Labels
Y_train = np.array([
[["Retriever", "Golden Retriever"], ["Hound", "Dachshund"]],
[["Retriever", "Labrador"]],
[["Hound", "Dachshund"], ["Hound", "Beagle"]],
], dtype=object)
# Use decision tree classifiers for every node
tree = DecisionTreeClassifier()
classifier = MultiLabelLocalClassifierPerNode(local_classifier=tree)
# Train local classifier per node
classifier.fit(X_train, Y_train)
# Predict
predictions = classifier.predict(X_test)
print(predictions)
Total running time of the script: ( 0 minutes 0.047 seconds)